- PhD Tutorial 2017
- BioPPN 2016
- Beijing 2015
- BioPPN 2015
- BioPPN 2014
- Petri Nets 2014
- BioPPN 2013
- Petri Nets 2013
- BioPPN 2012
- ICSB 2011
- BioPPN 2011
- AWPN 2010
- ISS - BioPN 2010
- BioPPN 2010
- Petri Nets 2009
- APBC 2009
- ISMB 2008
- BioSysBio 2008
data structures and software dependability
computer science department
brandenburg university of technology cottbus - senftenberg
T U T O R I A L - Petri nets for multiscale systems biology
latest update: March 08, 2019, at 12:53 PM
Basic facts
- satellite event of PETRI NETS 2013
- where: Milano / Italy
- when: Tuesday, June 25, 2013
- tutorial level: intermediate/advanced
- length: full day
- related event: BioPPN workshop, Monday, June 24, 2013
Index
- organizers/presenters
- abstract
- expected audience
- tools
- outline
- slides
- supplementary material and useful links
- assignments
- references
- registration
o r g a n i s e r s / p r e s e n t e r s
- David Gilbert - Brunel University, School of Information Systems, Computing and Mathematics, Uxbridge/London, UK
- Monika Heiner - Brandenburg University of Technology at Cottbus, Computer Science Institute, Germany
- Wolfgang Marwan - Otto von Guericke University Magdeburg, Departement of Regulatory Biology, Germany
supported by
- Mary Ann Blaetke, Otto von Guericke University Magdeburg, Germany
- Daniele Maccagnola, University Milano-Bicocca, Italy
- Ovidiu Parvu, Brunel University, UK
a b s t r a c t
The use of models of biochemical networks is a central component for both Systems and Synthetic Biology. Constructing, analysing and applying these models for prediction (Systems Biology) or design (Synthetic Biology) is a major challenge that can benefit from the application of methods originating in computer science and software engineering. This tutorial gives an introduction to a general modelling framework and shows how it can be applied to analysing existing biological systems and designing novel systems. A particularly challenging aspect is modeling biological systems which are characterized by important features at multiple spatial and/or temporal scales.
We introduce a biomodel engineering framework to address some of these issues within the context of multiscale Systems Biology. Our methodology is based on a structured family of Petri net classes which enables the investigation of a given system using various modelling abstractions: qualitative, stochastic, continuous and hybrid, optionally in a spatial context. We illustrate our approach with case studies demonstrating hierarchical flattening, treatment of space, and hierarchical organisation of space.
Finally, we discuss two approaches of a systematic network construction - design modules first and then construct the networks by module composition, or automatically reconstruct the networks from time series data.
e x p e c t e d . a u d i e n c e

- The target audience includes research students as well as researchers.
- Suitable for computer scientists and engineers familiar with basic modelling approaches, as well as for biochemists with an understanding of biochemical net- works.
- The tutorial illustrates the special challenges, which come with this application area, and also indicate some open problems which need to be solved. So the tutorial might be of special interest to those looking for new challenges to their theoretical background.
t o o l s
The tutorial material was developed using the following tools.
- Snoopy - design and animation/simulation of qualitative/stochastic/continuous/hybrid Petri nets;
- Charlie - standard analysis techniques of Petri nets theory;
- Marcie - reachability analysis and exact/approximative/simulative model checking of CTL/CSRL/PLTLc;
- MC2 - simulative PLTLc model checker for discrete/continuous traces.
o u t l i n e
1. Basics & background
- Biomodel engineering introduction
- Petri nets for biology
- The Petri net framework (4 x 2 classes, coloured & uncoloured)
- Petri nets – standard techniques and beyond
Break
2. Colour for space, levels & movement
- Encoding 1, 2, 3D
- Diffusion
- Growth
- Geometries
- Illustrative example: bacterial colony;
- Levels/multiscale,
- Illustrative examples: PCP or simpler
- Movement & history
Lunch
3. Simulation and analysis
- Simulation & unfolding
- Analysis techniques
- Model checking in the three basic paradigms
- Image analysis
Break
4. Systematic network construction
- Modular modelling - modules & model composition
- Algorithmic mutation
- Automatic network reconstruction
s l i d e s
1. Basics & background
2. Colour for space, levels & movement
3. Simulation and analysis
4. 4. Systematic network construction
s u p p l e m e n t a r y . m a t e r i a l . a n d . u s e f u l . l i n k s
- Petri nets for multiscale Systems Biology - online project summary
- beginner's guide - where to start reading
- Petri Nets in Systems Biology - tutorial book
a s s i g n m e n t s
To earn 1 credit point for taking part in the advanced tutorial requires to solve one of the assignments with a total work load of 30 hours. Please contact one of the organisers of this workshop if you are interested.
r e f e r e n c e s
- MA Blätke, A Dittrich, C Rohr, M Heiner, F Schaper and W Marwan:
JAK/STAT signalling - an executable model assembled from molecule-centred modules demonstrating a module-oriented database concept for systems and synthetic biology;
Molecular BioSystem, 2013. - MA Blätke, M Heiner and W Marwan:
Predicting Phenotype from Genotype Through Automatically Composed Petri Nets;
Proc. CMSB 2012, London, Springer, LNCS/LNBI 7605, pp. 87–106, 2012. - Q Gao, D Gilbert, M Heiner, F Liu, D Maccagnola and D Tree:
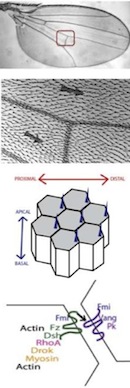
Multiscale Modelling and Analysis of Planar Cell Polarity in the Drosophila Wing;
IEEE/ACM Transactions on Computational Biology and Bioinformatics, 99(PrePrints):1-1, 2012. - D Gilbert, M Heiner, F Liu and N Saunders:
Colouring Space - A Coloured Framework for Spatial Modelling in Systems Biology;
Proc. PETRI NETS 2013, Milano, Springer, LNCS 7927, pp 230–249, June 2013. - M Heiner, D Gilbert and R Donaldson:
Petri Nets for Systems and Synthetic Biology;
SFM 2008, Springer, LNCS 5016, pp. 215–264, 2008. - M Heiner and D Gilbert:
How Might Petri Nets Enhance Your Systems Biology Toolkit;
Proc. PETRI NETS 2011, Springer, LNCS 6709, pages 17–37, 2011. - M Heiner and D Gilbert:
BioModel Engineering for Multiscale Systems Biology;
Progress in Biophysics and Molecular Biology, 111(2-3):119–128, April 2013 - W Marwan, C Rohr and M Heiner:
Petri nets in Snoopy: A unifying framework for the graphical display, computational modelling, and simulation of bacterial regulatory networks;
In Methods in Molecular Biology – Bacterial Molecular Networks, (Jv Helden, A Toussaint and D Thieffry, Eds.), Humana Press, pages 409–437, 2012. - O Parvu, D Gilbert, M Heiner, F Liu and N Saunders:
Modelling and Analysis of Phase Variation in Bacterial Colony Growth;
Proc. CMSB 2013, Vienna, Springer, LNCS, to appear, September 2013.
r e g i s t r a t i o n
Registration is handled by the hosting conference website PETRI NET 2013
q u e s t i o n s
Please contact one of the organisers.